Stem and Leaf Rot of Tomato Caused by Boeremia linicola
Article information
Trans Abstract
In December 2020, stem and leaf rot symptoms in small-fruited tomato (Solanum lycopersicum) plants were observed in a farmer's vinyl greenhouse located in Pyeongtaek, Gyeonggi Province, Korea. The incidence of diseased plants in the vinyl greenhouse was 2–6%. Seven single-spore isolates of Phoma sp. were obtained from the diseased stems and leaves. All the isolates were identified as Boeremia linicola based on the cultural, morphological and molecular characteristics. Two isolates of B. linicola were tested for pathogenicity on stems and leaves of small-fruited tomato and large-fruited tomato using artificial inoculation. All the tested isolates caused stem and leaf rot symptoms in the inoculated plants. The symptoms were similar to those observed in plants from the vinyl greenhouse investigated. This is the first report of B. linicola causing stem and leaf rot in tomato.
Tomato (Solanum lycopersicum) is native to Peru and widely cultivated as an annual crop in many regions of the world (Jones et al., 2014; Plants of the World Online, 2022). Tomato fruits are used as a popular vegetable and food. In Korea, farmers generally grow tomato in vinyl greenhouses. In December 2020, stem and leaf rot symptoms in small-fruited tomato plants were observed in a farmer's vinyl greenhouse located in Pyeongtaek, Gyeonggi Province, Korea. The symptoms on stems initially appeared as small, grayish black circular to fusiform spots. As the disease progressed, the lesions extended to 10 to 20 mm in length (Fig. 1A). The symptoms on leaves appeared as circular to irregular black spots in diameter of 5–10 mm. Later, the lesions expanded to more than 10 mm in diameter and were sometimes torn (Fig. 1B). Three sites were observed in the vinyl greenhouse, and 50 plants at each site were investigated for the disease incidence. The incidence of diseased plants in the vinyl greenhouse was 2–6%.
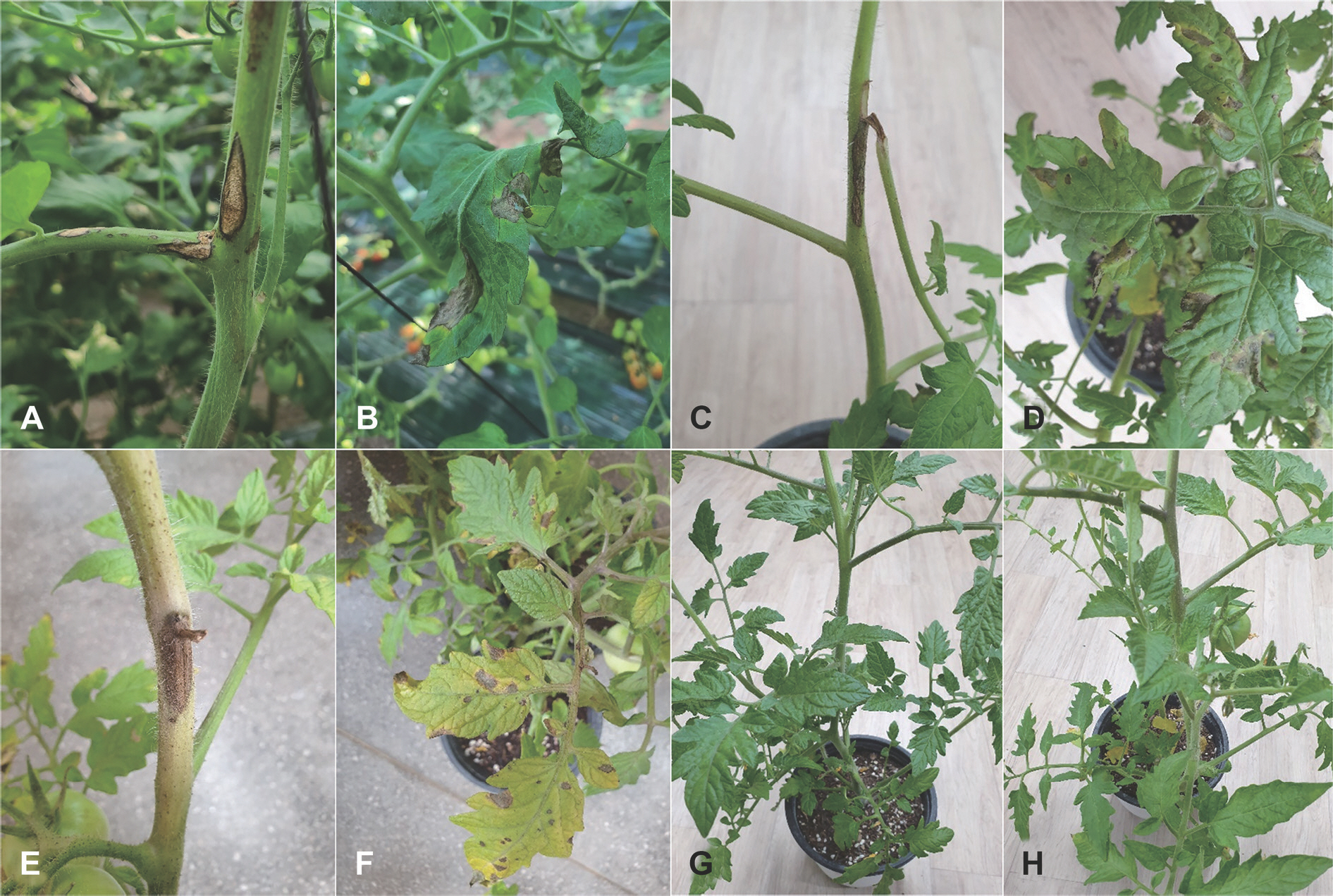
Stem and leaf rot symptoms of tomato plants. (A, B) Symptoms observed in small-fruited tomato plants in the vinyl greenhouse investigated. Symptoms induced by artificial inoculation tests with Boeremia linicola isolates in a small-fruited variety (C, D) and a large-fruited variety (E, F). Non-inoculated plants (control) of a small-fruited variety (G) and a large-fruited variety (H).
Diseased stems and leaves of tomato were collected from the vinyl greenhouse investigated, and fungal pathogen was isolated from lesions of the diseased stems and leaves. The 3–5 mm-long lesion pieces cut from the lesions were plated on 2% water agar (WA) after surface-sterilizing with 1% sodium hypochlorite solution for 1 min. The fungal mycelia growing from the lesion pieces were transferred to potato dextrose agar (PDA) slants after incubating the plates at 25 °C for 2–3 days. Morphological characteristics of the isolates cultured in PDA slants for 3–4 weeks were examined by a compound microscope (Nikon Eclipse Ci-L, Tokyo, Japan). Most of the isolates were identified as Phoma sp. based on the morphological characteristics described in a previous study (Boerema et al., 2004). The Phoma sp. isolates were transferred to oatmeal agar (OA) plates and incubated at 22 °C for 2–3 weeks. Conidial suspension of each isolate was prepared from the OA plate cultures and streaked on WA plate using a sterile loop. After incubation of the WA plate for 24 hr at 22° C, germinated conidia were picked up under a dissecting microscope (Nikon SMZ 1780) and transferred to new WA plates. Seven single-spore isolates of Phoma sp. were obtained from the WA plate cultures and cultured in PDA slants. The isolates were used for identification and pathogenicity tests.
The single-spore isolates of Phoma sp. were cultured on malt extract agar (MEA), OA, and PDA at 22° C for 14 days for investigation of their cultural and morphological characteristics using the methods described in previous studies (Boerema et al., 2004; Chen et al., 2015). Five culture plates of each isolate were used for each medium. Average diameters of 7-day-old cultures of the isolates grown on MEA, OA, and PDA at 22° C under darkness were 3.6 cm, 4.7 cm, and 3.7 cm, respectively. Colony morphology of the isolates on the three media was investigated after an additional incubation at 22 °C of the cultures in alternating cycles of 13 hr NUV light and 11 hr darkness for 7 days. The colony on MEA showed white to pale olivaceous velvety aerial mycelia (Fig. 2A). The colony on OA showed olivaceous gray to dark floccose aerial mycelia (Fig. 2B). The colony on PDA showed olivaceous gray velvety aerial mycelia (Fig. 2C). NaOH spot tests (Boerema et al., 2004) of the isolates on 7-day-old MEA cultures showed positive reactions (pigment α to β) (Fig. 2D). Morphological features of 10 pycnidia and 30 conidia per each isolate produced in three to 4-week-old OA cultures were examined by the compound microscope. Pycnidia were mainly globose with non-papillate to 1–2 papillate ostioles, solitary or confluent, usually submerged in the agar (Fig. 2E), and measured 66–210 μ m in diameter. Conidia were mainly aseptate, rarely 1-sepate, ellipsoidal to oblong, usually with small multi-guttules (Fig. 2F), and measured 3.0–11.2×2.0–4.4 μ m (av. 5.7×2.7 μ m). All the isolates were identified as Phoma exigua var. linicola based on the cultural and morphological characteristics described in previous studies (Boerema et al., 2004; Maas, 1965). P. exigua var. linicola was reclassified as Boeremia exigua var. linicola based on phylogenetic analysis (Aveskamp et al., 2010), and later as B. linicola (Jayawardena et al., 2019).

Cultural and morphological features of Phoma sp. isolate from tomato. Colonies of the isolate grown on malt extract agar (A), oatmeal agar (B), and potato dextrose agar (C) at 22° C for 14 days. (D) Reaction of NaOH spot test of the isolate on 7-day-old malt extract agar culture. A pycnidium of the isolate produced in oatmeal agar (E) and conidia produced in the pycnidium (F).
To verify the identification of P. exigua var. linicola isolates based on the cultural and morphological characteristics, three target genes (ITS, TUB2, and RPB2) of the isolates were investigated using the methods described in a previous study (Chen et al., 2015). Genomic DNA of the isolates was extracted using molecular grade Chelex 100 resin (Bio-Rad, Hercules, CA, USA) according to the protocol used by Blount et al. (2016), with slight modifications. Following primers for polymerase chain reaction (PCR) amplification of the genomic DNA were used with V9G (De Hoog and Gerrits van den Ende, 1998) and ITS4 (White et al., 1990) for ITS, TUB2Fd and TUB4Rd (Woudenberg et al., 2009) for TUB2, and RPB2-5F2 (Sung et al., 2007) and fRPB2-7cR (Liu et al., 1999) for RPB2. Conditions of PCR amplification for all the genes were followed as those discribed by Chen et al. (2015). All alignment works were performed using Bioedit version 7.2.5 (Hall, 2013). All PCR products were confirmed with 100 bp plus DNA ladder (Bioneer, Daejeon, Korea). Sequencing of genomic DNA was carried out based on the same primer sets at Bionics (Seoul, Korea). To construct a phylogenetic tree of the concatenated sequences with the three target genes, a maximum likelihood (ML) method was used. The ML method was conducted with 1,000 bootstrap replicates, which were performed by raxmlGUI version 2.0 (Edler et al., 2021) based on a general time reversible substitution model with inverse gamma-distributed rate variation (GTR+G+I). The final concatenated alignment included 10 ingroup taxa with a total of 1,472 characters including gaps (515 for ITS, 347 for TUB2, 610 for RPB2). Leptosphaeria doliolum (CBS 505.75) was set as the outgroup taxon. All the isolates were clustered in a group with B. linicola (synonym: P. linicola) CBS 116.76 (Fig. 3). The nucleotide sequences of ITS, TUB2, and RPB2 genes obtained from the seven isolates were deposited in GenBank with accession numbers of OL854118– OL854124, OL862983– OL862989, and OM716913– OM716919 for ITS, TUB2, and RPB2 genes, respectively.

Phylogenetic tree based on the concatenated sequences of ITS, TUB2, and RPB2 of seven Boeremia linicola isolates (TOPH-2001– TOPH-2020) from tomato and reference species. The reference sequence data were obtained from the NCBI GenBank data-base. The tree was generated using maximum likelihood method based on GTR+G+I model by raxmlGUI version 2.0. The bootstrap support values are given at the nodes. The bar represents the num-ber of nucleotide substitutions per site. Ex-type strains are marked by an asterisk (∗).
Two isolates of B. linicola from tomato were tested to confirm their pathogenicity on stems and leaves of two tomato varieties (Bunong Seed Corporation, Suwon, Korea), BN20-B104 (small-fruited variety) and BN21-A609 (large-fruited variety), using artificial inoculation. A conidial suspension (1–2×106 conidia/ml) of each isolate was harvested from 3-week-old cultures on OA. The conidial suspension was sprayed onto 2-month-old tomato plants grown in plastic pots (height, 15 cm; upper diameter, 17 cm; lower diameter, 10 cm) in a vinyl greenhouse. Twenty-five milliliters of conidial suspension of each isolate was sprayed onto each tomato plant. Control plants were treated with same quantity of sterile distilled water. Inoculated plants were placed in plastic boxes (80×59×48 cm) under 100% relative humidity at room temperature (24–26° C) for 5 days. After that, the inoculated plants were taken out of the plastic boxes and placed in a vinyl greenhouse. Ten days after inoculation, pathogenicity of the isolates was rated based on the degree of stem and leaf rot symptoms. The pathogenicity test was conducted in triplicate. All the tested isolates caused stem and leaf rot symptoms in the inoculated plants of two tomato varieties (Fig. 1C–F), but no symptoms occurred in the control plants (Fig. 1G, H). The lesions were more produced on the leaves than on the stems (Table 1). The symptoms of the inoculated plants were similar to those observed in plants from the farmer's vinyl greenhouse investigated. The inoculated isolates were reisolated from the lesions.
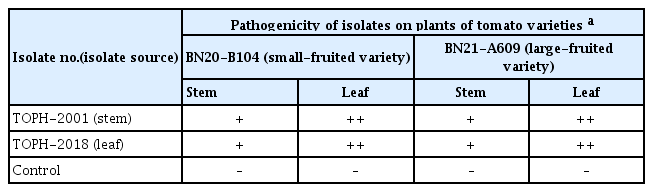
Result of pathogenicity tests of Boeremia linicola isolates on tomato plants by artificial inoculation
It has been reported that B. linicola (synonym: B. exigua var. linicola; P. exigua var. linicola) causes various diseases in Linium catharticum, L. usitatissium, Nemophila insignis, Salvia greggii, and Sesamum indicum (Farr and Rossman, 2022). Didymella stem rot caused by Didymella lycopersici (anamorph: Phoma lycopersici) and Phoma rot caused by Phoma destructiva var. destructiva were reported to occur in tomato (Jones et al., 2014). However, there has been no report on occurrence of stem and leaf rot in tomato caused by B. linicola. This is the first report of B. linicola causing stem and leaf rot in tomato.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
