Isolation and Characterization of Bacteriophages Infecting Ralstonia solanacearum from Potato Fields
Article information
Abstract
Bacterial wilt caused by Ralstonia solanacearum is one of the most devastating diseases in major Solanaceae crops. The pathogen is easily disseminated and survives for many years in plant farming system. Although chemicals are applied to control the disease, they are of limited efficacy and cause several problems. Therefore, the use of phage therapy has been suggested to control the disease as a biological agent. In this study, we discovered bacteriophages lysing diverse Ralstonia isolates from plant and soil samples obtained from the potato cultivated field in Jeju. Three times repeated pickings of plaques resulted in obtaining 173 single phages showing diverse spectrum of host-specificity. With the results, 12 core phages were selected and dendrogram was generated. Genetic diversity of the selected phages was also confirmed by AFLP (Amplified Fragment of Length Polymorphism) fingerprinting. The stability of the phages was investigated in various temperatures and various conditions of pH in vitro. The phages were stable at 16°C–44°C and pH 6–10. Morphological characterization of the phages revealed they were all classified into the Podoviridae, but had diverse head sizes. The results of this research will contribute to control the disease and further researches regarding genetic and molecular aspects will facilitate understanding phage and bacteria interaction.
Introduction
The phytopathogen, Ralstonia solanacearum is a soil-borne gram-negative bacterium and causes bacterial wilt disease in many important crops in tropical and subtropical areas (Hayward, 2000). However, the reports of the disease have increased in temperate regions including Republic of Korea due to the increased temperature caused by global warming (Shin and Yun, 2010). R. solanacearum is highly heterogeneous and was subdivided into five races and six biovars based on their physiological and biochemical characteristics (Hayward, 2000; Hayward et al., 1990). Recent classification revealed that it is sub-grouped into four phylotypes based on molecular and geographical information (Fegan and Prior, 2005; Remenant et al., 2010). Although this pathogen is easily disseminated and survives for many years in plant farming system and chemicals are applied to control the disease, they are of limited efficacy and cause several problems such as environment pollution, ecosystem disruption and residual chemicals on crops. Due to these reasons, the use of phage therapy has been suggested to control bacterial diseases as a biological agent and bacteriophage (phage) infecting R. solanacearum was also proposed to control bacterial wilt disease (Bae et al., 2012; Balogh et al., 2010; Fujiwara et al., 2011).
Bacteriophage is one of the most abundant organisms in the biosphere including rhizosphere and effective on bacterial evolution (Ashelford et al., 2003; Suttle, 2005). Phage is virus specifically infecting bacteria and its host specificity is very high. However, it is not toxic to any of eukaryotic cells including animals and plants and its preparation is easy and inexpensive (Ackermann et al., 1997; Jones et al., 2007). Bacteria can be lysed by infecting with a virulent phage that result numerous progeny phages released and neighboring bacteria can be continuously infected. Therefore, bacterial lysis amplifying phages results the minimization and control of bacterial diseases.
In spite of the huge diversity of phages, the investigation for the isolation and characterization of various phages infecting R. solanacearum to control is not sufficient in Korea. In this study, therefore, we isolated various phages infecting diverse and different range of R. solanacearum isolates from plants and soils collected from several potato cultivated fields in Jeju, Korea and characterized the selected phages.
Materials and Methods
Collection of experimental samples and Ralstonia isolates.
Samples used to isolate bacteriophages were collected from diverse potato fields in Jeju, Korea (Table 1). Six samples of diseased plant showing bacterial wilt symptom and six samples of soil where bacterial wilt habitually occurred were included. In total, 33 isolates of R. solanacearum were kindly provided by National Agrobiodiversity Center in Republic of Korea (Table 2). Of them, 14 and 19 were isolated from potato and other horticultural crops, respectively.

Infectivity of the 12 selected phages against 33 different isolates of Ralstonia solanacearum identified by the dotting assay
Isolation and selection of bacteriophages
Each plant and soil sample were homogenized with sterile water and centrifuged. Initially, supernatants obtained from each sample were mixed and cultured with each 14 potato-origin isolate of R. solanacearum in Nutrient Broth (NB; BD and Company, Sparks, MD, USA) medium at 28°C for 20 hours. After the overnight culture, the cultured samples were centrifuged and filtered through 0.45 μm pore-size filters (GVS, Bologna, Italy). These filtrates were applied in a plaque assay (double-layer agar method) against the 14 isolates to confirm the presence of bacteriophages. When plaques were observed, individual plant and soil samples were reacted with certain isolates and plaque assays were applied again to investigate isolate-spectrums of each plant and soil sample. Bacteriophages obtained from the initial isolation were purified by three times repeated picking of plaques and eluted in SM buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4) and they were stored at 4°C for further experiments. In each picking step, picked plaque (phage) samples were screened to investigate isolate-specificity with all 33 isolates of R. solanacearum by a dotting assay. Dendrogram was generated using the NTSys software (Exeter Software, Setauket, NY, USA) according to the isolate-specificities and plaque types ranged from 0 to 5 assigned depending on the clearance of plaques. The selection of bacteriophages showing diverse responses was performed considering the results of dendrogram in each picking step.
Purification of bacteriophages
The selected bacteriophages were propagated with certain isolates of R. solanacearum and bacteriophage lysates were obtained. The lysates were precipitated by treating with 10% polyethylene glycol (PEG) and purified using CsCl density-gradient ultracentrifugation at 30,000 rpm and 4°C for 3 hours. The purified bacteriophage particles were dialyzed and stored at 4°C.
DNA isolation and DNA fingerprinting of bacteriophages
Genomic DNAs of the selected and purified phages were isolated using the Phage DNA Isolation Kit (Norgen Biotek, Thorold, ON, Canada). To obtain a high concentrated phage DNA, we used a high density of phage lysates (>1á108 pfu/ml). The procedure was carried out according to the manufacturer’s instruction. To investigate DNA fingerprints of the selected phages, an AFLP (Amplified Fragment of Length Polymorphism) method was applied (Vos et al., 1995). Primary templates were prepared by using two restriction enzymes, PstI and MseI, ligated with adaptors fitting to the PstI and MseI sites, and 10 times diluted prior to the pre-amplification with primers fitting to both adaptors. For the selective amplification, four primer combinations, PA/MAAC, PA/MATC, PC/MCAC and PC/MCGA, coded by Vos et al. (1995) and van Os et al. (2006) were used. AFLP bands were separated on a 6% polyacryl amide gel and the gel was visualized using the Silverstar Staining System (Bioneer, Daejeon, Korea). Polymorphic bands were scored as 1 or 0 for presence or absence and the scored data was used to generate a phylogenetic tree using the NTSys software.
Characterization of bacteriophages
To investigate thermal stability of bacteriophage, phage aliquots from four randomly selected phages were incubated at various temperatures ranged from 16°C to 60°C for 15 minutes in three replications. Phage stability under various pH was also investigated in three replications. Phage aliquots from four randomly selected phages were mixed with diverse pH buffers ranged from 3 to 11 and incubated at 4°C for 3 hours. Initial concentration of the phages used for the investigation of both thermal and pH stabilities was approximately 100–200 pfu/ml. After incubation, the concentration of the phages was determined using the plaque assay and the rates of phage viability were calculated. The obtained data were statistically analyzed using an ANOVA and Duncan’s multiple range test at P<0.05. In addition to these, morphology of phages was assessed by transmission electron microscopy. Solution containing phage was placed on carbon-coated grid and 2% uranyl acetate was applied to the grids for 20 seconds before scanning images. Phages were classified according to the guidelines suggested in International Committee on Taxonomy of Viruses (Fauquet et al., 2005).
Results and Discussion
Isolation and selection of bacteriophages
In order to isolate phages infecting R. solanacearum, diseased plants and soils were collected from the potato cultivated fields where the symptom of bacterial wilt disease was observed in Jeju (Table 1). Initially, the samples were screened with the 14 potato-origin isolates of R. solanacearum that have diverse races and biovars (data not shown). Although all the samples were collected from the potato cultivated field in Jeju, host-specificities were diverse. Four and three from plants and soils samples showed responses to some of isolates, respectively, but the samples (P1 and S1, P3 and S3) collected from the same fields and another soil sample (S5) didn’t produce any plaque. P2 and P6 showed relatively broad host range, but S6 produced plaques with only one isolate. These results indicated that the phages can survive at both plant and soil and are naturally active at and/or in plants in the field. The characteristics of plaques detected were also diverse at the size and concentration of plaques (data not shown). These kinds of diversification such as different host-specificities and morphology of plaques at the isolated phages infecting R. solanacearum from the different samples have been reported by a few researchers (Askora et al., 2009; Fujiwara et al., 2008; Kawasaki et al., 2007; Murugaiyan et al., 2011; Yamada et al., 2007). This feature was also observed at the interaction between different bacterial species and phages in Korea (Jee et al., 2012).
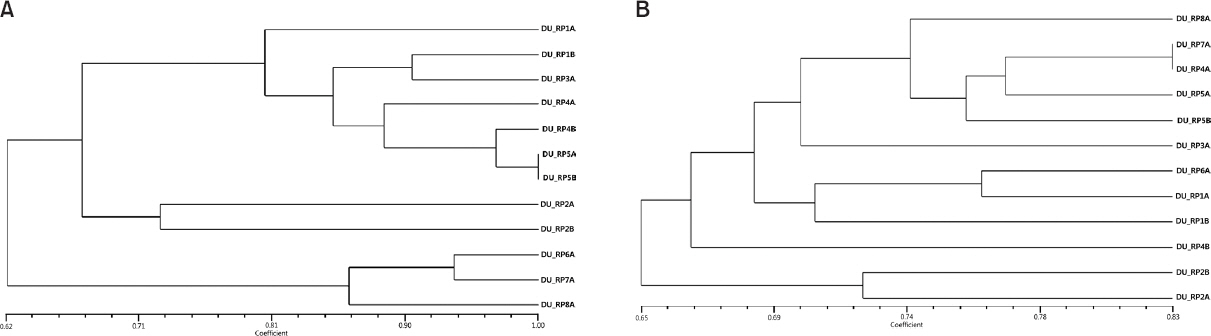
To purify the phages, single plaque pickings were performed and screening for the identification of host-specificity with the picked phages was expanded to the 33 isolates of R. solanacearum. In every picking step, host-specificity was investigated and plaque types were assigned to 0 to 5 depending on the clearance of plaques. The scored data allowed the dendrogram to be generated and core phages were selected for further researches. Finally, 173 single plaques were picked in total and 12 core single phages were selected for further characterization according to the results of host-specificity. They were named and numbered with DU_RP (Daegu University_Ralstonia Phage). DU_RP1A, 1B and 3A derived from P6, DU_RP2A, 2B, 4A and 4B derived from P2, DU_RP5A and 5B derived from P4, DU_RP6A derived from P5, DU_RP7A derived S2 and DU_RP8A derived from S4 (Table 2).
Host specificity of bacteriophages
Host-specificity of the 12 selected and purified phages was examined with the 33 isolates of R. solanacearum. All selected phages could lyse some of certain isolates tested (Table 2). DU_RP1A to 5B showed relatively broad-spectrum host range, but DU_RP6A to 8A showed relatively narrow host range. All of them could lyse 14 of 33 isolates of R. solanacearum, but patterns of lysis were slightly different each other. None of them infected 10 isolates and nine isolates could specifically be lysed by certain selected phages (Table 2). The dendrogram was generated based on host-specificity (Fig. 1A). The 12 selected phages were classified as three different groups. DU_RP5A and DU_RP5B had exactly same host range, but morphology of the two phages was different. They had very different head sizes (data not shown). In addition to these, phages separated from the same samples often showed different spectrum to the isolates. Some of the selected phages showed cell lysis to certain isolate although it could not lyse the isolate during initial screening in the original sample, and vice versa. It might be caused by the fact that some of phages were aggressive in the mixture of the phage sample and certain phages were discarded during the repeated picking and selection. This feature was found in our contemporary research with Pectobacterium species.
AFLP fingerprinting and phylogenetic analysis
In order to investigate genetic diversity of the selected phages, AFLP analysis was performed. In total, 126 polymorphic bands were scored from the selective amplification with four primer combinations, generating the phylogenetic tree (Fig. 1B). It confirmed that the 12 selected phages have different genomic composition and could result diverse host-specificity of the selected phages. However, when the phylogenetic tree was compared with the dendrogram generated based on the host-specificity, it was not completely fit to each other. A few selected phages were equally grouped in both trees. It might be caused by the fact that the data generated by AFLP derived from partial genomic regions.
Temperature and pH stabilities of bacteriophages
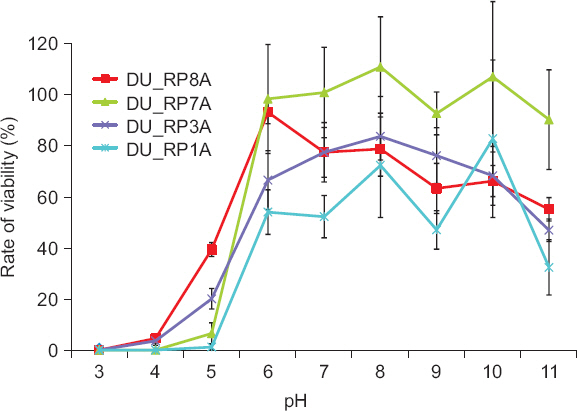
The temperature and pH stabilities of the phages were investigated. Four of the randomly selected phages were incubated at 16°C–60°C for 15 minutes and statistical analysis revealed that the phages were stable at 16°C–44°C (Fig. 2). However, phage stability decreased dramatically from 48°C and the phages were mostly inactivated at 56°C and 60°C. Depending on the selected phages, stability was significantly different (data not shown). Regarding pH stability, four of the randomly selected phages were also stored at pH 3–11 for 3 hours and the phages were statistically stable at pH 6–10 (Fig. 3). At pH 11, the phage stability was slightly reduced, but the phage was almost or completely inactivated at pH 3–5. The viability of phages was significantly different depending on the selected phage samples indicating that aggressiveness of phages on bacterial cell lysis could be different. The results of thermal and pH stabilities obtained in this study were slightly different from those reported by Bae et al. (2012) and Lim et al. (2013). The Podoviridae phage for lysing Ralstonia was stable in a range of temperature from 15°C to 60°C and pH from 4 to 11 (Bae et al., 2012) and the Podoviridae phage for lysing Pectobacterium species was stable in a range of temperature from 20°C to 40°C and pH from 4 to 11 (Lim et al., 2013). The phages selected in this study were slightly more sensitive to environmental conditions. This might be caused due to different environments which phages were isolated from. Overall, the results obtained in this study suggested that the tested phages were stable at a wide range of temperature, but could not be stable at strongly acidic soil condition. This should be considered when the phages were applied as a biocontrol agent in the field.
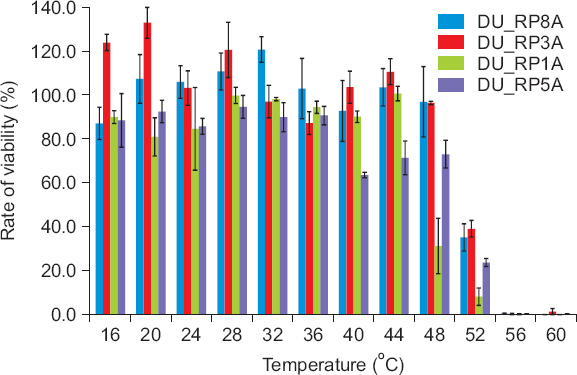
Stability of bacteriophages at various temperatures. Phage lysate aliquots were incubated at different temperatures for 15 minutes.
Morphological characteristics of bacteriophages
The selected phages were propagated and purified using the PEG and CsCl density-gradient ultracentrifugation methods. The purified phage particles were subjected to electron microscopic observation. The images of the 12 phages revealed that they have diverse head sizes ranged approximately from 40 to 110 nm in diameter and very short tail and they were classified into the Podoviridae family in the order Caudovirales. The images of two phage particles are present in Fig. 4. Previously a Podoviridae phage infecting R. solanacearum isolated from a pepper rhizosphere has been reported by Bae et al. (2012) in Korea, but the phage could be different from some of the phages identified in this study due to smaller or bigger head sizes and different origin. Several Podoviridae phages have been isolated in other countries as well (Ackermann, 2003; Bhunchoth et al., 2015; Kawasaki et al., 2009), but there are no evidence for difference and similarity. Therefore, further characterization should be acquired. A few filamentous phages infecting R. solanacearum have also been reported (Kawasaki et al., 2007; Murugaiyan et al., 2011; Yamada et al., 2007).
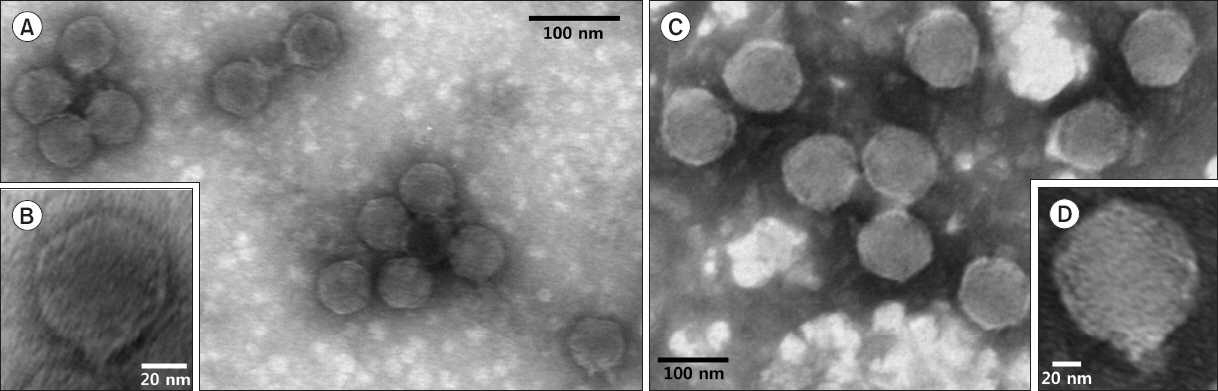
The transmission electron microscopic images of two selected phages. (A, B) DU_RP1B. Podoviridae with small size of head and very short tail. (C, D) DU_RP1A. Podoviridae with large size of head and very short tail.
This is the first report of R. solanacearum-infecting phages isolated from potato plants and potato rhizosphere in Korea. The results obtained in this research will contribute to control the disease and further researches regarding genetic and molecular aspects will facilitate understanding phage and bacteria interaction. Further characterization of the phages and application of the phages in potato cultivation has still been in progress.
Acknowledgement
This study was supported by the fund of cooperative research program for National Institute of Crop Science (Project number: PJ0 11190032016), Rural Department Administration, Republic of Korea.
References
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.