Development of a Multiplex PCR for Simultaneous Detection of Blueberry Red Ringspot Virus and Blueberry Scorch Virus Including an Internal Control
Article information
Abstract
Blueberry red ringspot virus (BRRSV) and blueberry scorch virus (BlScV) are included in the quarantine virus list managed by the Korean Animal and Plant Quarantine Agency. A multiplex polymerase chain reaction (PCR) assay with an internal control was developed for the simultaneous detection of both viruses. The specific primers used here were designed based on the highly conserved regions of the genomic sequences of each virus, obtained from the National Center for Biotechnology Information nucleotide databases. The primers were designed to amplify a partial sequence within coat protein (CP) for detecting BRRSV and a partial sequence within the CP-16 kDa for detecting BlScV. 18S ribosomal RNA (rRNA) was used as internal control, and the primer set used in a previous study was modified in this study for detecting 18S rRNA. Each conventional PCR using the BRRSV, BlScV, and 18S rRNA primers exhibited a sensitivity of approximately 1 fg plasmid DNA. The multiplex PCR assay using the BRRSV, BlScV, and 18S rRNA primers was effective in simultaneously detecting the two viruses and 18S rRNA with a sensitivity of 1 fg plasmid DNA, similar to that of conventional PCR assays. The multiplex PCR assay developed in this study was performed using 14 blueberry cultivars grown in South Korea. BRRSV and BlScV were not detected, but 18S rRNA was all detected in all the plants tested. Therefore, our optimized multiplex PCR assay could simultaneously detect the two viruses and 18S rRNA in field samples collected from South Korea in a time-efficient manner. This approach could be valuable in crop protection and plant quarantine management.
Blueberry is a member of the genus Vaccinium of the family Ericaceae, which is native to North America. Blueberry is widely consumed worldwide and is considered a super-food, owing to its potent antioxidant activity and immune-boosting qualities (Banerjee et al., 2020). In South Korea, the number of imported and cultivated blueberries is increasing annually (Song et al., 2014).
Presently blueberries have been cultivated in several countries, increasing the risk of complex viral outbreaks. The viruses that infect blueberries are as follows: blueberry green mosaic-associated virus (BGMaV), blueberry latent spherical virus (BLSV), blueberry latent virus (BBLV), blueberry leaf mottle virus (BLMV), blueberry mosaic associated virus (BlMaV), blueberry necrotic ring blotch virus (BNRBV), blueberry red ringspot virus (BRRSV), blueberry scorch virus (BlScV), blueberry shock virus (BlShV), blueberry shoestring virus (BBSSV), blueberry virus A (BVA), blueberry virus T (BlVT), cherry leaf roll virus (CLRV), peach rosette mosaic virus (PRMV), strawberry latent ringspot virus (SLRSV), tobacco ringspot virus (TRSV), and tomato ringspot virus (ToRSV) (Saad et al., 2021).
Among blueberry-infecting viruses, BRRSV belonging to the genus Soymovirus (family Caulimoviridae) has a circular double-stranded DNA genome (Fauquet et al., 2005). BRRSV-infected blueberries exhibit red ringspots on both the leaves and stems. These ringspots are circular and light-colored blotches on fruits; additionally, these fruits are often deformed and exhibit delayed ripening (Isogai et al., 2009). Since this viral disease was first observed in the highbush blueberry in New Jersey in the 1950s, it has expanded to Japan (2009), Slovenia (2009), the Czech Republic (2010), Poland (2011), and Serbia (2012), as well as other states in the United States (1995) (Isogai et al., 2009; Jevremović et al., 2015; Martin et al., 2012; Petrzik et al., 2011). In South Korea, BBRSV was first reported in a highbush blueberry cultivar Duke cultivated in Pyeongtaek in 2012 (Cho et al., 2012).
BlScV belonging to the genus Carlavirus (family Betaflexiviridae) has a positive-sense single-stranded RNA genome (Saad et al., 2021). BlScV-infected blueberries exhibit full blighting of blossoms, chlorosis, red line shape of young leaves, and stem dieback (Saad et al., 2021). The yield in susceptible cultivars is reduced significantly (Bristow et al., 2000). Since this virus was first reported in the United States in the 1980s, it has been discovered in Canada (2004), Germany (2013), Italy (2005), The Netherlands (2008), Poland (2013), and China (2018) (Ciuffo et al., 2005; Kalinowska et al., 2013; Saad et al., 2021; Wegener et al., 2004; Xie et al., 2018).
BRRSV and BlScV have been frequently reported in blueberries cultivated in several of the aforementioned countries mentioned; hence, the viruses are included in plant quarantine requirements for blueberries imported into South Korea (Plant Quarantine Technology Center in Animal, Plant and Fisheries Quarantine and Inspection Agency, 2013). However, we lack efficient techniques for their detection that can prevent possible outbreaks in the fields.
In this study, we developed a multiplex PCR assay with 18S rRNA as an internal control for simultaneous detection of the two viruses to improve the reliability of detection. We believe that our study would facilitate crop protection and quarantining of imported plants.
Specific primers for detecting two viruses (BRRSV and BlScV) were designed based on the highly conserved regions of their genomic sequences (Table 1). The genomic sequences of the two viruses were obtained from the National Center for Biotechnology Information (NCBI) nucleotide databases (BRRSV: NC_003138, HM159264, JF421559, MZ004932, JN205460; BlScV: NC_003499, AY941198, KP232980, KP232983, KP232986, KP232990, and KP232997). Primers were designed to amplify partial sequences within the partial coat protein (CP) of the BBRSV genome and the partial CP-16 kDa of the BlScV genome. A pair of 18S ribosomal RNA (rRNA) primers used in our previous study (Park et al., 2021) was modified to use it for all blueberry plants assessed in this study and as an internal control for multiplex PCR assay. The 18S rRNA gene has been used as an internal control for detecting DNA and RNA viruses in host plants (Abade Dos Santos et al., 2021; Dayaram et al., 2012; Park et al., 2021; Tang and Lightner, 2000). The expected sizes of the products amplified by the BRRSV, BlScV, and 18S rRNA primers in the monoplex PCR assays were 601, 357, and 855 bp, respectively (Table 1).
The sensitivity of the monoplex and multiplex PCR assays was evaluated using a 10-fold dilution series of an equal mixture of the three recombinant plasmid DNAs. The recombinant plasmid DNAs were obtained using the pGEM-T Easy Vector (Promega, Madison, WI, USA). Each recombinant plasmid DNA contained the respective amplicon of the two viruses and 18S rRNA (601, 357, and 855 bp). They were used as a positive control template in the monoplex and multiplex PCR assays. Amplicons of the two viruses were synthesized according to their genomic sequences. The monoplex PCR assays were performed in a 50 µl reaction volume containing 5 µl of plasmid template, 1× buffer without MgCl2 (TaKaRa, Tokyo, Japan), 1.25 U Taq DNA polymerase (TaKaRa), specific primers (upstream and downstream primers), and 4 µl of dNTPs (2.5 mM). Each reaction contained 5 µl of serially diluted plasmid template with amounts ranging from 10 ng to 1 fg per reaction. The recommended primer concentrations for monoplex PCR were 1,000 nM (BRRSV), 100 nM (BlScV), and 100 nM (18S rRNA) for each primer. Magnesium and bovine serum albumin (BSA) have been used as enhancers for PCR (Chang and Mahoney, 1995; Özay and McCalla, 2021); therefore, magnesium and BSA were used to optimize PCR efficiency in this study. Optimal PCR efficiency was achieved using a final concentration of 6 mM MgCl2 (TaKaRa) and 0.004% BSA (TaKaRa). The monoplex PCR reactions were conducted in a SimpliAmp Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) using an initial denaturation step at 95 o C for 3 min, followed by 40 cycles of denaturation at 95 o C for 30 sec, annealing at 60 o C for 30 sec, and extension at 72 o C for 30 sec, and a final extension at 72 o C for 10 min. All the resulting PCR amplicons were analyzed using electrophoresis on 1% agarose gels with a 1 kb plus DNA ladder (Thermo Fisher Scientific). The results of the monoplex PCR assays revealed that BRRSV, BlScV, and 18S rRNA produced the desired amplicons of 601, 357, and 855 bp, respectively. The specific primers for BRRSV and BlScV designed in this study could amplify the respective plasmid DNA with a sensitivity approaching 1 fg (Fig. 1). The specific primers for 18S rRNA, available as an internal control, also exhibited similar sensitivity (Fig. 1). The multiplex PCR assay was performed under the same conditions (i.e., as a monoplex PCR). However, it was performed using three plasmid templates. The resulting detection limit was 1 fg for each plasmid DNA (Fig. 2).
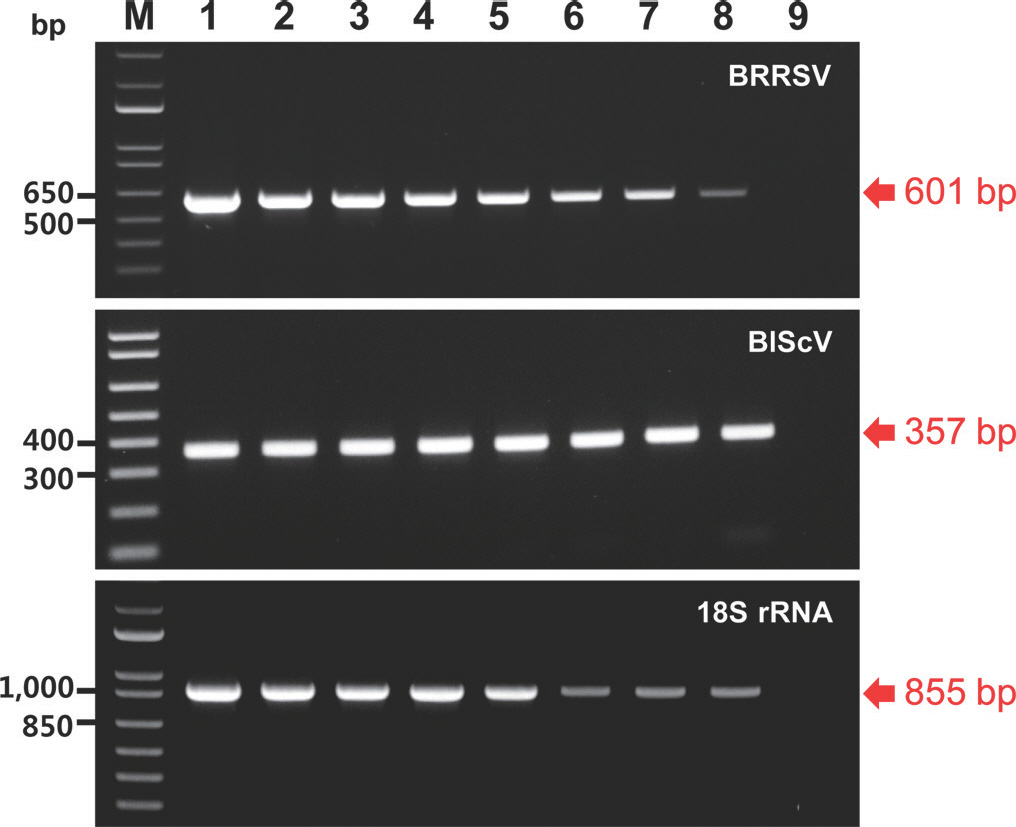
Agarose gel analysis of products obtained using the monoplex polymerase chain reaction (PCR) assays of 10-fold serial dilutions of the corresponding recombinant plasmids carrying the indicated amplicons. M, 1 kb plus DNA ladder; lanes 1 to 8, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg of plasmid, respectively; lane 9, negative control (empty vector); BRRSV, blueberry red ringspot virus; BlScV, blueberry scorch virus. Red arrows indicate the location of PCR amplicons of the expected sizes.
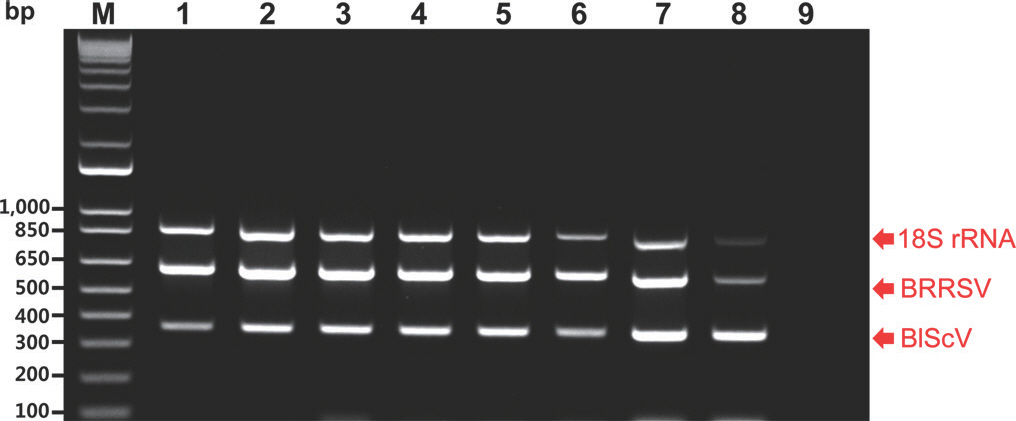
Agarose gel analysis of products obtained using the multiplex polymerase chain reaction (PCR) assays of the two viruses using 10-fold serial dilutions of an equal mixture of the three different recombinant plasmids, including the 18S rRNA plasmid (18S). M, 1 kb plus DNA ladder; lanes 1 to 8, products from template mixtures containing 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg of each plasmid, respectively; lane 9, negative control (empty vector); BRRSV, blueberry red ringspot virus; BlScV, blueberry scorch virus. Red arrows indicate the location of PCR amplicons of the expected two viruses and 18S rRNA.
To apply the monoplex and multiplex PCR assays to field samples, the samples were collected from 14 blueberry (Vaccinium corymbosum) cultivars grown in South Korea as follows: ‘Chandler’, ‘Chanticleer’, ‘Draper’, ‘Duke’, ‘Elizabeth’, ‘Georgia dawn’, ‘Huron’, ‘Mae’, ‘Mega blue’, ‘New Hanover’, ‘O'Neal’, ‘Reka’, ‘Sierra’, and ‘Suziblue’. Total DNA for detecting BRRSV was isolated from the leaves of 14 blueberry cultivars using the cetyltrimethylammonium bromide (CTAB) method. The CTAB buffer used in this study was a solution containing 2% CTAB, 0.1 M Tris-HCl (pH 8.0), 20 mM EDTA (pH 8.0), and 1.4 M NaCl. Total DNA isolated from 14 blueberry cultivars was used in the monoplex PCR with specific primers for detecting BRRSV and 18S rRNA primers as internal controls. The monoplex PCR assay was performed under the same conditions described earlier. PCR products of the expected size for BRRSV were not detected, whereas those of 18S rRNA was detected in the monoplex PCR assay (Fig. 3A).
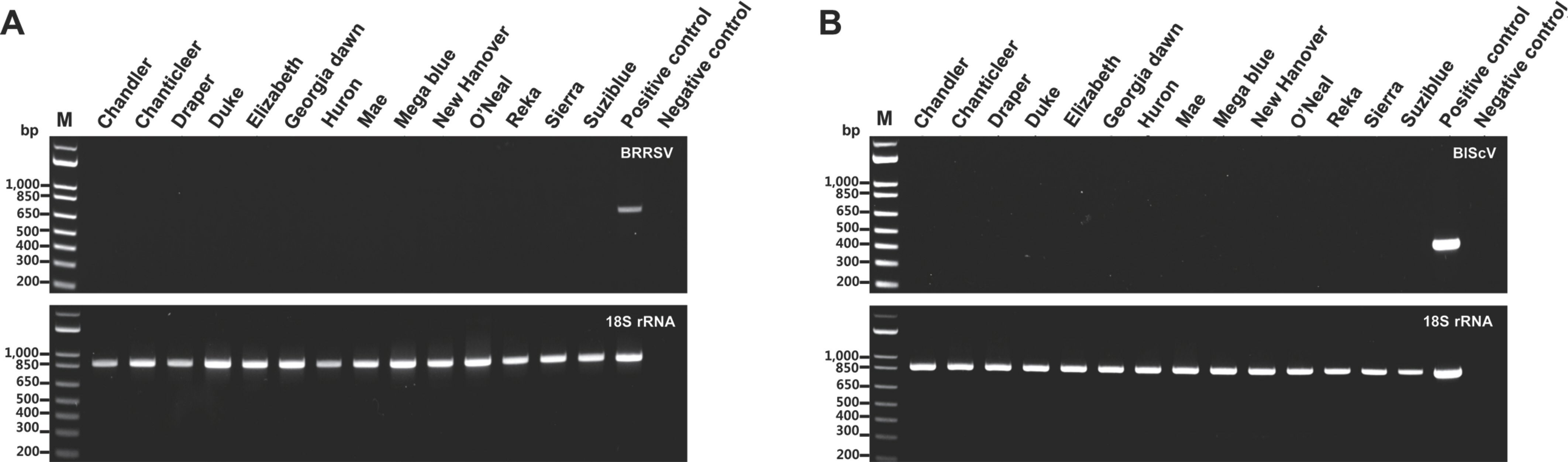
Monoplex polymerase chain reaction (PCR) assays for the two viruses in 14 blueberry (Vaccinium corymbosum) cultivars. The PCR products to detect blueberry red ringspot virus (BRRSV) (A) and blueberry scorch virus (BlScV) (B), including 18S used as internal control, are displayed. M, 1 kb plus DNA ladder; positive control (each target plasmid); negative control (empty vector).
Total RNA for detecting BlScV was isolated from the leaves of 14 blueberry cultivars using the Total RNA Prep Kit (BioFact, Daejeon, Korea). The total RNA obtained in this study was used in the reverse transcription (RT) reaction for cDNA synthesis. The RT reaction was performed in a 50 µl reaction volume containing 1 µg of total RNA, 1× buffer (Thermo Fisher Scientific), 100 U RevertAid reverse transcriptase (Thermo Fisher Scientific), random hexamer primers, and 4 µl of dNTPs (2.5 mM). The PCR machine for the RT reaction was the same as described earlier, and the reaction was conducted at 45 o C for 1 hr and then at 72 o C for 10 min. cDNA was synthesized as described earlier. The synthesized cDNAs were used in monoplex PCR with specific primers for detecting BlScV and 18S rRNA primers as internal controls. Monoplex PCR for BlScV detection was performed under the same conditions described above. The targeted PCR amplicons for BlScV were not detected, whereas those for 18S rRNA were detected in the monoplex PCR assay (Fig. 3B).
To detect DNA and RNA viruses simultaneously, total nucleic acids were isolated from the leaves of 14 blueberry cultivars using the CTAB method described by Chang et al. (2007). CTAB-based methods for the simultaneous detection of RNA and DNA viruses have been used in various plants (Abarshi et al., 2012; Chang et al., 2007; Henderson and Hammond, 2013). The total nucleic acids obtained in this study were used in multiplex PCR with specific primers for detecting BBRSV and BlScV and the 18S rRNA primers as internal controls. The multiplex PCR assay was performed under the same conditions as described earlier. PCR products of the expected size for the two viruses were not detected. In contrast, those of 18S rRNA were detected in the multiplex PCR assay (Fig. 4). Consequently, BRRSV and BlScV were not detected in any of the blueberry cultivars examined in this study.
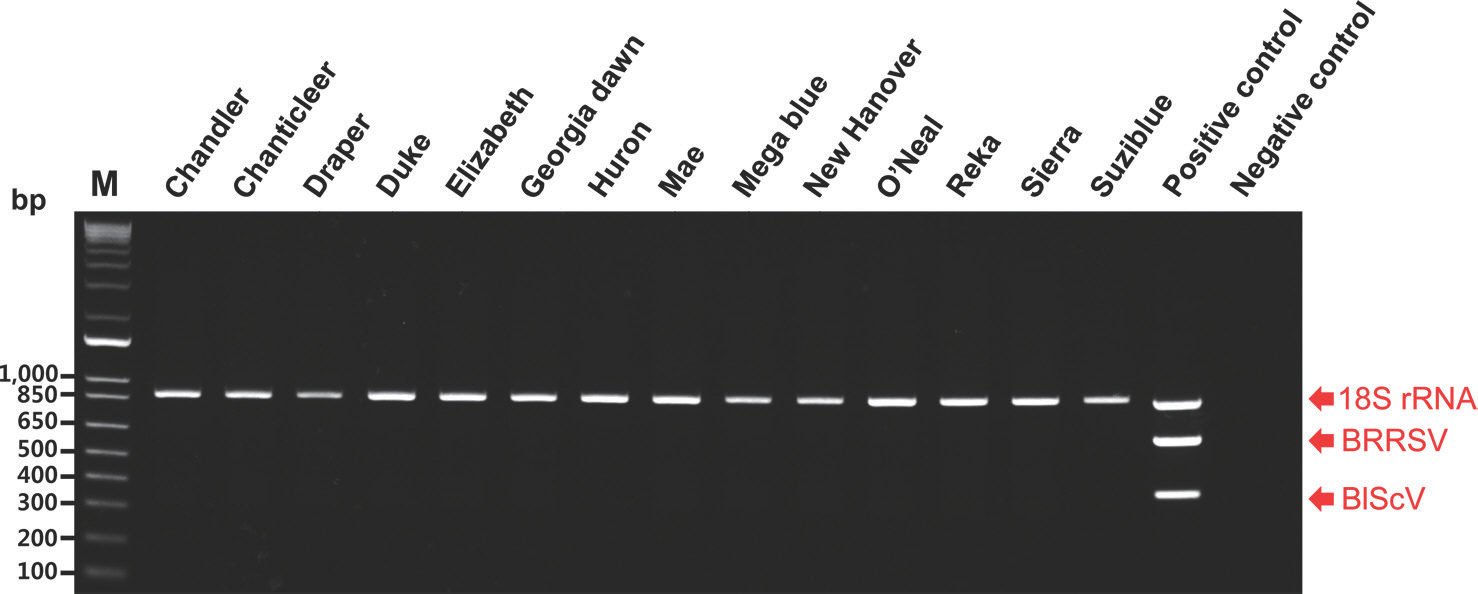
Multipelx polymerase chain reaction assay for the simultaneous detection of the two viruses in 14 blueberry (Vaccinium corymbosum) cultivars. M, 1 kb plus DNA ladder; positive control (a mix of three plasmid templates); negative control (empty vectors); BRRSV, blueberry red ringspot virus; BlScV, blueberry scorch virus.
In a previous study, a multiplex PCR assay for virus detection in blueberry plants was developed based on the ToRSV and TRSV (Fuchs et al., 2010); however, multiplex PCR for the detection of BRRSV and BlScV had not been developed. The detection limits of multiplex PCR capable of detecting ToRSV and TRSV were not mentioned by Fuchs et al. (2010); therefore, our study may be the first to explicitly report that two viruses (BRRSV and BlScV), including internal control, can be detected simultaneously in multiplex PCR.
In conclusion, we developed a multiplex PCR assay that simultaneously detects RNA and DNA viruses capable of infecting blueberry plants. This assay can be used for crop protection and contexts linked to the quarantining of plants.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This study was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Crop Viruses and Pests Response Industry Technology Development Program, funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (grant number 320044-3). This study was also supported by a sab-batical year research grant from Seoul Women's University (2021).
