Occurrence of Damping-off Caused by Pythium spinosum on Cucumis melo in Korea
Article information
Abstract
In 2010 and 2012, damping-off symptoms were found on melon seedlings grown in Yeongam and Suncheon, Korea. Water-soaked and discolored lesions appeared on the lower stems of the infected plants. The diseased plants became wilted and stunted, and eventually collapsed. On the basis of morphological characteristics and molecular analyses of internal transcribed spacer (ITS) rDNA and cytochrome oxidase II (cox2) sequences, the causal organism was identified as Pythium spinosum. The isolates were pathogenic to melon under pot conditions. To our knowledge, this is the first report of P. spinosum causing damping-off on melon in Korea.
Body
Melon (Cucumis melo L.) is an economically important horticultural crop grown throughout the world. Global export of melons produced in Korea has increased in recent years (http://www.kati.net). The cultivated area of melon increased by 13.1% to 1,500 ha in 2014 compared with 2004 (MAFRA, 2015). In 2010 and 2012, damping-off symptoms were observed on melon seedlings in greenhouses in Yeongam and Suncheon. Approximately 5% to 10% of seedlings showed the symptoms. The affected stems and roots were water-soaked, shriveled, softened, and discolored (Fig. 1A). Longitudinal cracks appeared on the lower parts of some stems (Fig. 1B). In the late stage of the disease, the infected plants collapsed and eventually decayed.

Damping-off caused by Pythium spinosum on Cucumis melo seedlings. (A) Symptoms of water-soaked and discolored lesions on the lower parts of stems. (B) A crack developing longitudinally at the base of the plant. (C, D) Similar symptoms occurred on seedlings after 10 days of inoculation.
A Pythium-like fungus was isolated from the diseased plants using the single hyphal tip method. A total of two pure isolates were obtained (Fig. 2A), and these were deposited in the Korean Agricultural Culture Collection under accession numbers KACC47889 and KACC47890 (Table 1). Morphological features were observed and measured from the isolates grown on water agar for 7 days using the grass blade culture technique. Primary hyphae were up to 7 µm in width, and hyphal swellings were often present (Fig. 2B). Oogonia containing plerotic oospores were globose, terminal or intercalary, and 18 to 21 µm in diameter, with spines on the oogonial walls (Fig. 2C, Fig. 2E). Antheridia were 1(–2) per oogonium, and mostly monoclinous (Fig. 2D). On the basis of the morphological data, the fungal pathogen was identified as Pythium spinosum Sawada (Van der Plaats-Niterink, 1981).
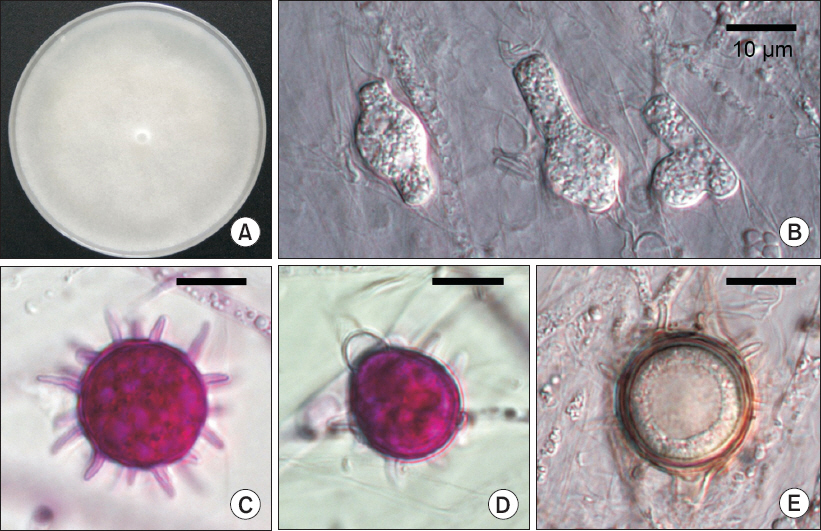
Cultural and morphological characteristics of Pythium spinosum from Cucumis melo. (A) Colony grown on potato dextrose agar after incubation for 5 days. (B) Hyphal swellings. (C) Oogonium ornamented with conical projections. (D) Oogonium with antheridium. (E) Oospore. Scale bars=10 µm (B–E).
Genomic DNA was extracted from mycelia scraped from agar plates. The internal transcribed spacer (ITS) rDNA region was amplified using the primers ITS1 (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3’) (White et al., 1990). The cytochrome oxidase II (cox2) gene was also amplified using the primers Cox2-F (5’-GGC AAA TGG GTT TTC AAG ATC C-3’) and Cox2-RC4 (5’-TGA TTW AYN CCA CAA ATT TCR CTA CAT TG-3’) (Choi et al., 2015). The PCR amplicons were purified and directly sequenced. The raw sequence data for P. spinosum were edited using SeqMan (DNASTAR Lasergene, Madison, WI, USA). The resulting sequences of the isolates have been lodged in GenBank, with accession numbers KT006894, KT006895, KT962996, and KT962997. To infer phylogenetic relationships between P. spinosum and related species, 12 reference sequences of Pythium spp. were retrieved from GenBank. Phytophthora parvispora (CBS 411.96) was used as an outgroup. Sequence alignment and phylogenetic analysis, using the neighbor-joining method with the Tajima-Nei model, were carried out in MEGA6 (Tamura et al., 2013). BLAST searches using the ITS rDNA and cox2 gene sequences of the present isolates identified high similarities (99%) with those of P. spinosum. In the phylogenetic tree (Fig. 3), the present isolates were grouped with a reference isolate of P. spinosum (CBS 275.67). Therefore, the molecular analyses verified the results of morphological identification.
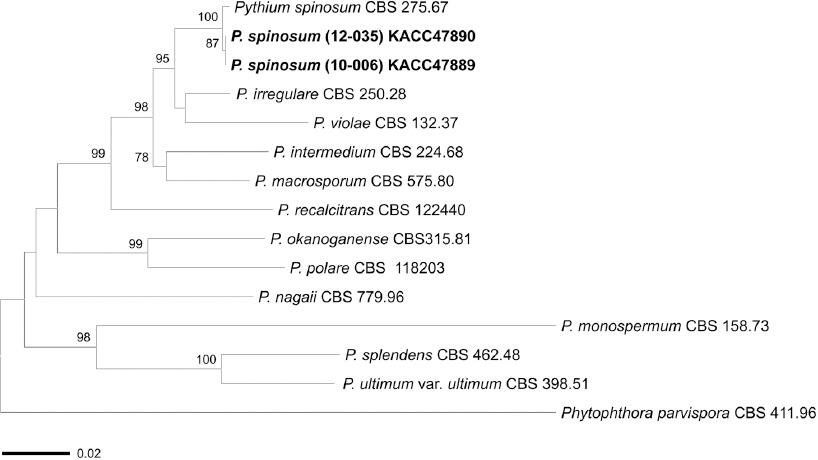
A neighbor-joining tree based on the combined internal transcribed spacer (ITS) rDNA and cox2 gene sequence datasets showing phylogenetic affinities of the two isolates of Pythium spinosum obtained in this study with isolates of other Pythium spp. Phytophthora parvispora was designated as the outgroup. Isolates in boldface were sequenced in this study. Bootstrap values above 70% are shown at the nodes. The scale bar represents 0.02 nucleotide substitutions per site.
The pathogenicity of the isolates was tested using 5-week-old seedlings of the melon cultivars Earthtalent, Asiaseongha, and Gyeouldaebak. Inoculum was prepared by harvesting mycelial mats from 7-day-old cultures grown on potato dextrose agar. Ten healthy plants of each cultivar were inoculated with hyphal suspensions, which were poured into each pot. Another ten plants sprayed with sterile water served as controls. After inoculation, the plants were maintained in a growth chamber at 23°C for 1 week. Infected plants showed typical symptoms of damping-off after 10 days of inoculation (Fig. 1C, Fig. 1D), whereas control plants remained symptomless. The identity of the fungus consistently re-isolated from the lesions after inoculation was confirmed by morphological examination and gene sequence analysis. The two isolates proved to be pathogenic to all three tested cultivars of melon, thereby fulfilling Koch’s postulates.
Several species of Pythium, e.g., P. aphanidermatum, P. debaryanum, P. spinosum, and P. ultimum, have been known to cause diseases such as damping-off and root rot on melons (Farr and Rossman, 2014). In particular, occurrences of damping-off caused by P. spinosum on melon have been previously recorded from Australia and Japan. In Korea, among melon pathogens, P. ultimum is associated to Pythium root rot, and Rhizoctonia solani is known to cause damping-off and stem rot on melon (KSPP, 2009). To our knowledge, this is the first report of damping-off associated with P. spinosum on melon seedlings in Korea. The disease has the potential to cause considerable yield loss of melons in greenhouses, particularly nurseries, not only in Korea but also throughout the world.
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgement
This work was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01136801)”, Rural Development Administration, Republic of Korea.