Draft Genome Sequence of a Chitinase-producing Biocontrol Bacterium Serratia sp. C-1
Article information
Abstract
The chitinase-producing bacterial strain C-1 is one of the key chitinase-producing biocontrol agents used for effective bioformulations for biological control. These bioformulations are mixed cultures of various chitinolytic bacteria. However, the precise identification, biocontrol activity, and the underlying mechanisms of the strain C-1 have not been investigated so far. Therefore, we evaluated in planta biocontrol efficacies of C-1 and determined the draft genome sequence of the strain in this study. The bacterial C-1 strain was identified as a novel Serratia sp. by a phylogenic analysis of its 16S rRNA sequence. The Serratia sp. C-1 bacterial cultures showed strong in planta biocontrol efficacies against some major phytopathogenic fungal diseases. The draft genome sequence of Serratia sp. C-1 indicated that the C-1 strain is a novel strain harboring a subset of genes that may be involved in its biocontrol activities.
body
Some plant-associated bacteria have been widely isolated and commercialized owing to their ability to inhibit the growth of plant pathogens and to promote plant health by various mechanisms (Kim et al., 2011b). Previously, we developed a chitin-based effective biocontrol formulation containing three chitinase producing-bacterial culture mixtures of Chromobacterium sp. C61, an unidentified strain C-1, and strain C-3, which has been used successfully to control Alternaria blight and anthracnose of ginseng (Kim et al., 2010), Phytophthora blight of pepper (Kim et al., 2008), and the growth of root-knot nematodes (Ha et al., 2014) in the field. The specific bacterial strains were selected due to their different antifungal activities against phytopathogenic fungi; for example, the strain C-1 strongly inhibited the growth of Phytophthora capsici, Chromobacterium sp. strain C61 was mainly antagonistic to Rhizoctonia solani, and the strain C-3 was antagonistic to R. solani and Fusarium spp. The draft genome sequence of the Chromobacterium sp. C61 has been determined and applied to understanding the biocontrol related traits (Kim et al., 2011a; 2014). In addition, a recent publication using the draft genome sequence of the C61 strain indicated that Chromobacterium sp. C61 produced a cyclic lipopeptide, chromobactomycin, and an extracellular chitinase, which contribute to its biocontrol activity (Kim et al., 2014). However, the genetic and biochemical trait(s) involved in the biocontrol activity of the strains C-1 and C-3 have not been understood so far. The specific objectives in this study were to evaluate the in planta biocontrol activity of the C-1 strain and to sequence and annotate the genome of C-1 strain to obtain insight into the mechanisms underlying its biocontrol activity.
Library construction and genome sequencing of C-1 were performed as described previously (McSpadden Gardner et al., 2014). Briefly, the genomic DNA of the C-1 isolate was isolated using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA). The library for Illumina sequencing was prepared from the DNA fraction of ~300 bp using Illumina paired-end sample preparation Kits (Illumina, San Diego, CA, USA). This library was sequenced in one lane of a standard flow cell on Illumina genome analyzer II for 76 cycles, generating ~6.3 million paired-end reads, each 75 nt long, which was equivalent to ~471 million nucleotides. The short-read sequences were assembled using Velvet version 0.7.55 (Zerbino and Birney, 2008; Zerbino et al., 2009) with an optimal hash length of 37 nucleotides and a minimum contig length of 150 nucleotides. The assemblies were uploaded to the automated annotation platform - Rapid Annotation using SubsystemsTechnology (RAST) server maintained by the National Microbial Pathogen Data Resource (Aziz et al., 2008) and visualized using the SEED viewer (Overbeek et al., 2005).
The assembled shotgun genome sequence of the C-1 strain with annotation was deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/genomes/wgs.html) under the accession numbers for 133 contigs CAQ001000001 to CAQO01000133 and the accession number for 70 scaffolds HG424324 to HG424393. This shotgun genome sequence of the C-1 has a total of 5,655,814 nucleotides with 56% G+C content. Annotation indicated that 70 scaffolds harbor 5,158 protein-encoding genes (PEGs), and 61 RNA sequences with 58 tRNA sequences were identified (Table 1). Most of PEGs were belonged in primary metabolisms, such DNA, RNA, protein, respiration, cell wall and capsule, amino acids and derivatives, and carbohydrates (Table 1). The subsystem category distribution analysis in the SEED viewer indicated that a few genes involved in secondary metabolism (3), dormancy and sporulation (3), and phages and transposable elements (10). Sequence coverage was 94-fold or greater for 99% of the annotated genes.
In the SEED viewer, the “sequence-based comparison” was used for the amino acid-sequence-based genome-wide comparison among the annotations obtained in this study. This series of comparisons was performed in a pair-wise manner where two annotations were compared each time, with one annotation chosen as the “reference organism” and the other chosen as the “comparison organism.” Each pair-wise genome-wide comparison revealed the amino acid sequence identities of all the protein encoding genes (PEGs) from the “reference organism” (reference PEGs) to those of their respective best PEG hits from the “comparison organism” (comparison PEGs).
Based on information from the public SEED database, the bacterial draft genome sequence of isolate C-1 was found to be most closely related to that of Serratia proteamaculans 568. The analysis indicated that 4,466 (87%) of the annotated PEGs in C-1 were found in the genome of S. proteamaculans 568 with an average sequence identity of 75%. Most of the unique 693 PEGs in C-1 showed hypothetic functions and phages or mobile elements (phage FluMu or prophage, Table 1). Genes encoding proteins possibly responsible for biocontrol potential were unique in C-1; some of these included genes encoding non-ribosomal peptide synthetase, phenazine biosynthesis protein PhzF, extracellular serine protease, and antibiotic biosynthesis monooxygenase. The sequence analysis indicated that at least five different genes encoding extracellular chitinase were identified in Serratia sp. C-1. In S. marcescens, prodigiosin (a red colored antimicrobial pigment) is known to be a key biocontrol compound (Harris et al., 2004; Li et al., 2015), but the genes encoding prodigiosin were not found in C-1. The absence of the prodigiosin-encoding gene in C-1 is supported the fact that C-1 did not produce any colored pigments (data not shown). Interestingly, the pvc gene clusters involved in the biosynthesis of a novel pyoverdine paerucumarin in Pseudomonas aeruginosa (Clarke-Pearson and Brady, 2008) and the genes involved in bacteriocin biosynthesis were found in the C-1 genome (Table 1). A recent report indicated that a S. marcescens Mm isolated from a pest-infected hazelnut plant produced a bacteriocin-like substance as an antibacterial agent (Ugras et al., 2014). We found 13 bacteriocin-related genes in the subsystem category distribution analysis of the C-1 genome (Table 1). We also found that the C-1 possesses strong antibacterial activity against various phytopathogenic bacterial species (data not shown). The genome sequence of the chitinase-producing C-1 strain will allow the study and identification of the important structural and regulating genes.
Because the ribosomal RNA genes could not be properly reconstructed during the draft genome assembly of C-1, the full 16S rRNA sequence was amplified as described previously (Han et al., 2012) using a PCR premix and a gradient thermo cycler (Bioneer Inc., Daejeon, Korea). The PCR products were purified using a PCP purification kit (Bioneer) and cloned into a pGEM T-easy vector (Promega, Madison, WI, USA). Sequencing of the PCR products was performed at Solgent Inc., (Daejeon, Korea) and the sequences were analyzed using BLAST. The full 16S rRNA sequence of the C-1 was fully reconstructed in the draft genome sequence. The results of the BLAST analysis indicated that the PCR product of C-1 showed the highest homology with Serratia species. Therefore, full-length 16S rRNA sequences of Serratia type strains were obtained from the public GenBank. Multiple alignment of the 16S rRNA sequence of Serratia sp. C-1, edited using BioEdit version 7.0, was conducted using MEGA version 6 based on the neighbor-joining method with the Kimura two-parameter model (Kimura, 1980) with bootstrap values based on 1,000 replications. Phylogenetic analyses of the 16S rRNA sequence indicated that C-1 is a novel strain of Serratia species (Fig. 1). The 16S rRNA sequence of Serratia sp. C-1 did not show high similarities with any type strains of Serratia species, especially the Serratia species used as biocontrol agents, such as S. plymutica and S. marcescens (De Vleesschauwer and Hofte, 2003; Harris et al., 2004).
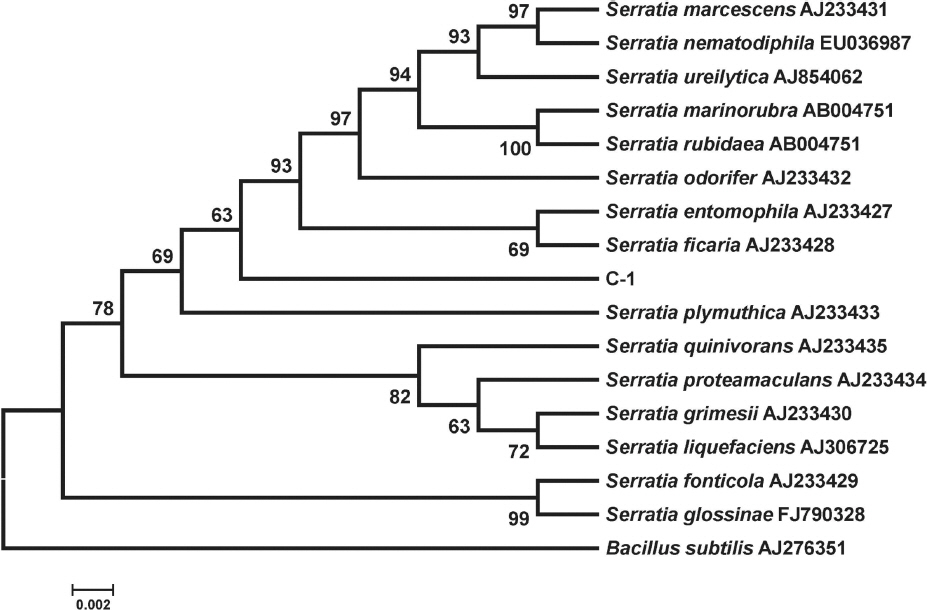
Phylogenetic tree of 16S rRNA sequences showing relationships between C-1 and the type strains of Serratia species constructed using the neighbor-joining method and Kimura’s two-parameter model. The numbers indicated at several nodes are consensus bootstrap values out of 1,000 replications. Scale bar, 0.002 substitutions per nucleotide position. Bacillus subtilis was used as the out-group. Accession numbers of the Serratia type strains are given at the end of the names of the strains.
The in planta biocontrol activity of C-1 was compared to that of Bacillus subtilis QST713, which is the main active ingredient of a commercial biocontrol product Serenade Max™ (AgraQuest Inc., CA, USA). C-1 and B. subtilis QST713 were grown in LB broth (Difco Inc., Detroit, MI, USA) for 3 days at room temperature with agitation in a shaking incubator. The bacterial cultures were diluted 3-fold with distilled water, and used to evaluate in planta biocontrol efficacies against major plant pathogens as described previously (Kim et al., 2014; Park et al., 2013). As negative controls, plants were treated with sterile water. After 24 h, the treated plant seedlings were inoculated with spores or mycelial suspensions of one of the plant pathogens as described previously (Kim et al., 2001). The experiment was conducted three times with 3 plants for each treatment, and the standard deviation was represented. Data were analyzed using IBM SPSS Statistics version 21 (IBM Corporation, Somers, NY, USA). The effect of the bacterial treatments on disease severity was determined using student’s t-test (P<0.01).
Foliar applications of C-1 culture supernatants resulted in significant decrease in the incidence of diseases such as rice sheath blight, tomato leaf blight, wheat leaf rust, and pepper anthracnose to an extent equal to or greater than that observed with B subtilis QST713 (Fig. 2).
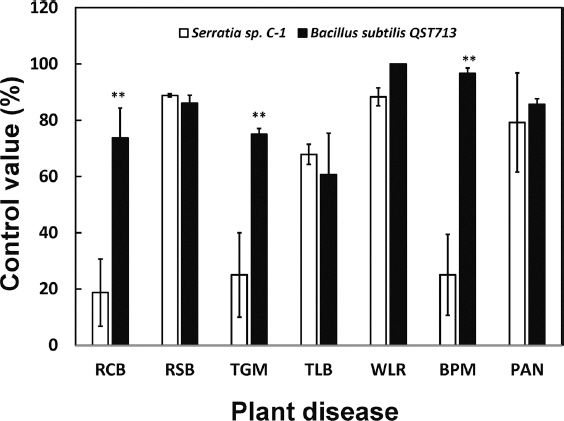
In planta biocontrol efficacy of the Serratia sp. C-1 bacterial cultures. Serratia sp. C-1 and Bacillus subtilis QST713 grown in LB broth at 25°C for 3 days with agitation at 150 rpm. Control plants were treated with sterile water. Data shown are averages and the standard error from three independent bioassays with three plants/treatment. Pathosystems evaluated include rice blast (RCB), rice sheath blight (RSB), tomato grey mold (TGB), tomato leaf blight (TLB), wheat leaf rust (WLR), barley powdery mildew (BPM), and pepper anthracnose (PAN). Asterisk (**) indicates significant differences in test values between treatments with Serratia sp. C-1 and B. subtilis QST713, as determined using student’s t-test (P<0.01).
In summary, we report that Serratia sp. C-1 bacterial cultures showed strong in planta biocontrol efficacies against some major phytopathogenic fungal diseases. The draft genome sequence indicated that Serratia sp. C-1 is a novel strain harboring a subset of genes that may be involved in its biocontrol activities. We are currently attempting the identification of genes possibly involved in the biocontrol activity and the regulation of their expression using the draft genome sequence determined in this study. We are also trying to study the biochemical nature of the metabolites of C-1 that can be important candidates for use in promoting plant health.
Acknowledgments
This work was supported by Chonnam National University, 2014.