Introduction
Plant viruses cause seriously economic losses in crops worldwide. Of plant viruses that infect tomato (Solanum lycopersicon), Tomato yellow leaf curl virus (TYLCV) is one of the most devastating viral diseases in tomato production. In most cases, commercial tomato plants infected with TYLCV at an early stage bear no fruits or malformation of fruits, causing losses up to 100% particularly in tropical and subtropical regions (Brown and Idris, 2008; Czosneck and Laterrot, 1997; Moriones and Navas-Castillo, 2000). Tomato flowers may appear but usually will drop before tomato fruits are formed. Tomato plants infected with TYLCV has severely reduced internodes of the plants, showing stunt symptoms. The new developing leaves in the infected tomato plants reduced greatly in size show wrinkled mottle symptoms and yellowing between the veins, and have margins that curl upward, giving them a cup-like appearance (Czosneck and Laterrot, 1997).
TYLCV is primarily transmitted by the sweet potato whitefly (Bemisia tabaci) and the silverleaf whitefly (Bemisia tabaci Biotype B = Bemisia argentifolii). These whitefly species are able to acquire TYLCV within 5 minutes by feeding on infected tomato plants, and the acquired whiteflies remain infective for life. Furthermore, TYLCV can be transmitted to the progenies of viruliferous whiteflies for at least two generations (Ghanim et al., 1998) and the virus is transmitted from viruliferous males of whiteflies to females and from viruliferous females to males but not among whiteflies of the same sex (Gahnim and Czosneck, 2000). TYLCV is not spread by other whitefly species such as the greenhouse whitefly (Trialeurodes vaporariorum) which is also common in plastic tunnels in farms, Korea. TYLCV cannot be spread by seeds and mechanical inoculation (e.g., by pruning equipment or by wound through touch). TYLCV is a member of the genus Begomovirus that belongs to the family Gemini-viridae. TYLCV genome consists of circular single-stranded DNA (ssDNA) molecules. One or two ssDNA molecules are present in one virus depending on the virus strain. Each ssDNA is approximately 2.5-2.8 Kb (Czosneck, 2007). TYLCV was first reported in Israel in 1964 (Cohen and Harpaz, 1964) and later observed in some states in USA in 1990s (Momol et al., 1999; Polston et al., 1999; Valverde et al., 2001). In addition, TYLCV was found in EU countries, Australia, Africa and Central America countries, causing seriously economic losses (Aidawati et al., 2002; Duffy and Holmes, 2007; Maruthi et al., 2005; Tsai et al., 2011a, 2011b). In Korea, TYLCV was observed in Tongyoung, 2008 and has been spread throughout the country (Lee et al., 2010).
Since 1998, a few serological detection methods based on polyclonal antibodies (PAbs) have been demonstrated to detect TYLCV from tomato. But, the PAbs developed were only efficient for western blot analysis rather than enzyme-linked immunosorbent assay (ELISA) in tomato samples collected from fields (Al-Bitar and Luisoni, 1995). Monoclonal antibodies (MAbs) using a recombinant TYLCV capsid protein (CP) expressed from E. coli used as an immunogen were efficient for TAS-EuSA assay against five begomoviruses including TYLCV (Wu et al., 2012). Recently, dot-EuSA and direct tissue blot immunoassay have been developed using six different MAbs against TYLCV, showing these methods were efficient for TYLCV detection in both field samples and insect vectors (Xie et al., 2013). Since these serological methods for TYLCV detection are indispensable to develop efficient antibodies, they require relatively long time, labors and technical skills. Meanwhile, molecular detection methods such as PCR analysis are widely used for TYLCV detection. In particular, PCR analysis is sufficiently sensitive and suitable for routine TYLCV diagnosis in tomato samples. However, clean DNA extraction is prerequisite to PCR analysis or other molecular methods for TYLCV detection.
In this study, we described a newly developed duplex PCR method for TYLCV diagnosis without DNA extraction. We showed that our duplex PCR method is a simple and sensitive TYLCV detection from tomato plants in a variety of farms, Korea.
Materials and Methods
Virus source and DNA extraction
Infectious DNA clones of 5 TYLCV isolates (for example, strain CW, TYLCV-CW) was used as source viruses (Choi et al., 2013). To obtain biologically active TYLCV strains, a single colony of Agrobacterium tumefaciens strain C58C1 containing a binary construct was inoculated to Luria-Bertani media supplemented with 10 mM MES (pH 5.6), 20 ŌĆöM acetosyringone (Fluka, USA) and the antibiotics (rifampicin 10 ╬╝g/ml, ampicillin 50 ╬╝g/ml and kanamycin 50 ╬╝g/ml). The cells were grown at 28┬░C overnight, and then the cells were collected by centrifugation. The precipitated cells were resuspended in infiltration buffer [10 mM MgCl2,10 mM MES (pH 5.6), and 150 ╬╝M acetosyringone] to a final OD600 of 1.0, as describe previously (Yoon et al., 2011). The first true leaf and cotyledons of tomato (cultivar Rokusanmaru) were agro-inoculated using the bacterial culture containing the DNA clones of TYLCV isolates.
Primers and PCR analysis
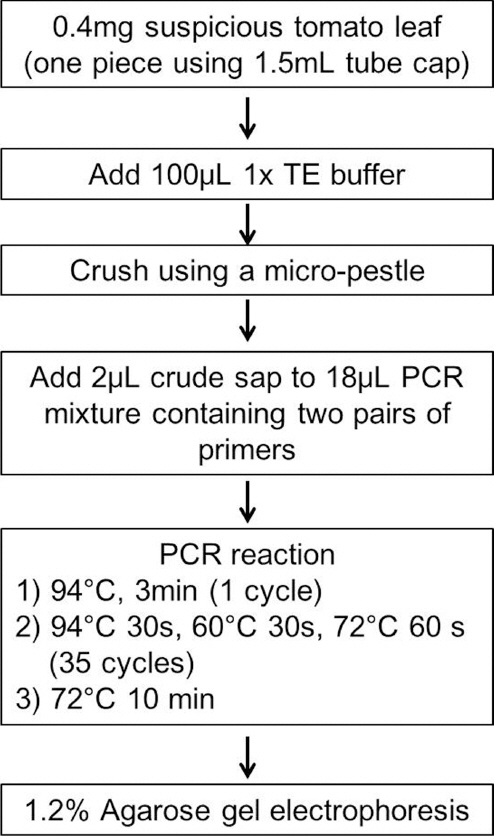
The prime pair TyCWF789 (5ŌĆÖ-tcgataatgagcccagtacc-3ŌĆÖ) and TyCWR1543 (5ŌĆÖ-aagcgtgtagacctagactg-3ŌĆÖ) was designed to amplify 754 bp TYLCV DNA fragment, based on the complete nucleotide sequences of TYLCV isolates deposited in the Genbank database. Accession information of TYLCV isolates analyzed in this study is provided on request. For internal control of PCR analysis, the primer pair To-TUBF857 (5ŌĆÖ-ctcagacctgaggaaattgg-3ŌĆÖ) and To-TUBR1325 (5ŌĆÖ-cattccttcacctgtgtacc-3ŌĆÖ) was designed to amplify 468 bp tomato ╬▓-tubulin DNA fragment. The nucleotide sequence of tomato ╬▓-tubulin gene (DQ205342) deposited in the Genbank database was used for design of internal primers. Approximately 0.01 g tomato leaf sample was extracted in 100 ╬╝l 1x TE buffer [10 mM Tris buffer (pH 8.0), 1 mM EDTA (pH 8.0)] using a plastic micro-pestle in a 1.5 ml tube. As a control, total DNA was extracted from tomato leaves infected with TYLCV-CW using DNeasy mini kit (Qiagen, USA). PCR amplification was carried out using a mixture of 20 ╬╝l PCR cocktail pre-mixture containing 10 mM Tris-HCl (pH 8.3), 2 mM MgCU 50 mM KCl, 10 pmol each of two pairs of primers (TyCWF789 + TyCWR1543 and To-TUBF857 + To-TUBR1325), 0.2 mM each dNTP, 1 unit of Hot-Start Taq DNA polymerase (Bioneer, Korea), and 2 ╬╝l supernatant of the extracted tomato sample. DNA fragments were amplified by PCR in a programmable DNA thermal cycler (BioRad, model C1000); 35 cycles (94 ┬░C 30 s, 60┬░C 30 s and 72┬░C 60 s) after an initial denaturation for 3 min at 95┬░C. In the 35th cycle, the final extension was for 10 min at 72┬░C.
Results and Discussion
To develop a simple and cost-effective PCR method for TYLCV detection, two novel pairs of primers were designed to amplify DNA fragments of TYLCV and tomato ╬▓-tubulin gene, respectively. This duplex PCR amplification has an advantage on checking of PCR error during PCR amplification for TYLCV detection. Firstly, we set up to determine the specificity and efficiency of designed duplex PCR method using newly designed primers for TYLCV detection. Total DNA was extracted from upper tomato leaves systemically agro-infected with TYLCV-CW and from healthy tomato leaves, according to manufacturerŌĆÖs instructions (Qiagen, USA). Duplex PCR reaction analyzed using the purified total DNAs showed that a single TYLCV DNA product of 754 bp and a single tomato ╬▓-tubulin DNA fragment of 468 bp were obtained from tomato samples infected with TYLCV-CW, while a single tomato ╬▓-tubulin DNA fragment of 468 bp was amplified from healthy tomato leaves (Fig. 1A). The expected sizes of PCR products were further confirmed using DNA sequencing analysis, indicating the expected sizes of PCR products was authentic (data not shown). Consistent duplex PCR amplification reactions were observed at the annealing temperature between 50┬░C and 60┬░C. None of PCR products was amplified from water as a negative control, suggesting that the two pairs of primers are highly specific to their target gene portions and the primers have no cross reactions in the tomato samples to detect TYLCV isolates.
Fig.┬Ā1
A duplex PCR detection analysis for TYLCV. (A) Total DNA was extracted from tomato leaves infected with TYLCV (Ty) and healthy tomato leaves (H), respectively. PCR was carried out in a final 20 ╬╝l volume, as described by Yoon et al. (2011) to determine specificity and sensitivity of the pairs of primers (see the text for details). PCR products were separated on 1.2% agarose gel and stained with edithium bromide solution. As a negative control, distilled water (H2O) was used to check nonspecific reactions in PCR amplification. A 100 bp DNA ladder (M) was used as a size marker. DNA size was indicated on the left. (B). Crude sap from tomato leaves infected with TYLCV (5 samples) and healthy tomato leaves (3 samples) were used for the duplex PCR reaction using the pairs of primers. To compare the duplex PCR reaction, total DNA of TYLCV-infected tomato (5 samples) and healthy tomato (5 samples) prepared from a commercial kit was subjected to the duplex PCR analysis. Total DNA from tomato leaf infected with TYLCV strain CW (Ty) and from a healthy tomato (H) was used as positive and negative controls. PCR products amplified were separated on 1.2% agarose gel and stained with ethidium bromide solution. DNA size was indicated on the left.
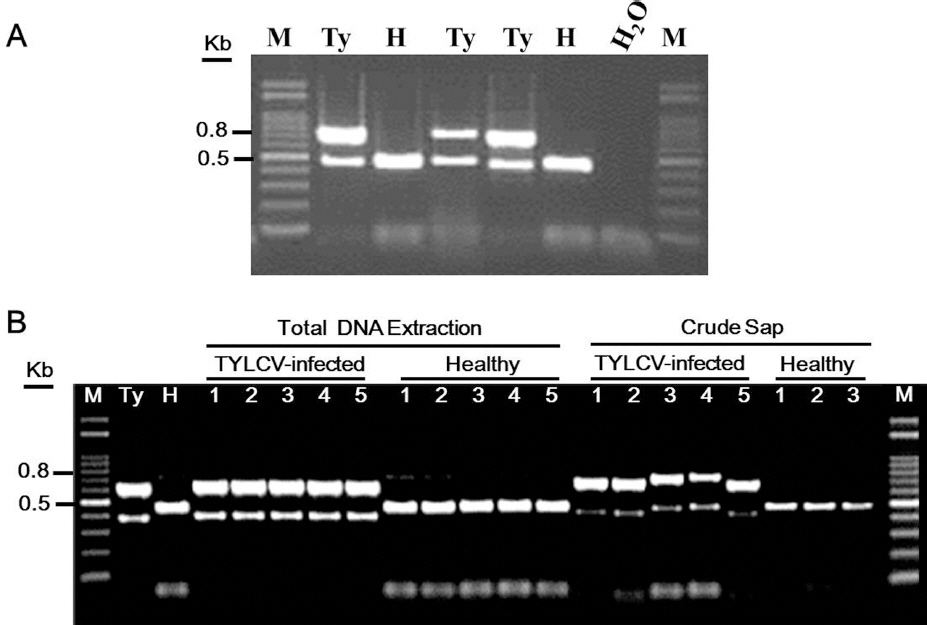
In principle, most of molecular diagnostic methods for plant viruses, including conventional PCR, require DNA extraction prior to TYLCV detections. Secondly, we were to determine the specificity and efficiency of designed duplex PCR method using newly designed primers without DNA extraction. To reduce time and labors during DNA extraction for TYLCV detection, crude sap without DNA extraction from each tomato leaf infected with 5 TYLCV isolates was subjected to duplex PCR amplification with two pairs of primers, respectively. Ten milligrams tomato leaf sample infected with TYLCV or the equivalent amount of healthy tomato leaf was crushed with 100 ╬╝l of 1x TE buffer using a micro-pestle in a 1.5 ml tube, respectively. This simple procedure requires 1 minute. Two micro-liters of crude sap were added to a mixture of PCR amplification in a final 20 pl volume. Two single PCR products were synthesized from the tomato samples infected with TYLCV isolates, as observed for duplex PCR amplification using total DNA (named Ty) from TYLCV-infected tomato (Fig. 1B). We found that centrifugation (for using supernatant) after crushing tomato leaf samples did not enhance the specificity and quality of our duplex PCR amplification (data not shown). PCR product for tomato ╬▓-tubulin gene, but not for TYLCV was only amplified from crude sap from healthy tomato leaves. Sizes of TYLCV PCR products from total DNA were the same like those of TYLCV PCR products from crude sap. The PCR products of tomato ╬▓-tubulin were observed in the size differences between total DNA and crude sap. Possible inhibition of PCR by tissue extract components was assumed. To clarify this issue, we determined whether the synthesized length of tomato ╬▓-tubulin products and TYLCV products from crude sap were different from those from total DNA using sequencing analyses. Sequence analysis of TYLCV PCR products and tomato ╬▓-tubulin PCR products showed no differences in length between sample extractions (data not shown), indicating that shift of PCR products during agarose gel electrophoresis might be caused by some salts and undesirable chemical reactions. As summarized in Fig. 2, these results suggest that newly developed duplex PCR method using crude sap from tomato samples infected with TYLCV is a simple, rapid and efficient diagnostic method, which the method allows saving of time and labors for TYLCV diagnosis.
To determine the detection limit and specificity of the developed duplex PCR method used directly from crude sap, various DNA concentrations were prepared from serial dilutions of total DNA and crude sap from tomato leaves infected with TYLCV. To use direct comparison for this duplex PCR method, we determined the amount of total DNA from 10 mg tomato leaves using a commercial DNA extraction kit. As results, we repeatedly obtained 1 mg total DNA from 10 mg crude sap of tomato plants infected with TYLCV (data not shown) when the concentration was determined using a DNA prep kit. As starting with 1 mg total DNA in PCR amplification, two single DNA products specific to the TYLCV gene and tomato ╬▓-tubulin gene were synthesized from tomato samples infected with TYLCV (Fig. 3A). Similar results were obtained from serially diluted crude saps from tomato leaves infected with TYLCV. The primers were able to synthesize successfully target DNAs in one zepto total DNA scale (named 10-15 in Fig. 3) in both batches (extracted total DNA and crude sap). These results suggest that our developed PCR method that can be used crude sap directly is high specificity for TYLCV detection in tomato plants.
Fig.┬Ā3
Determination of specificity and sensitivity of duplex PCR protocol for TYLCV diagnosis and application on field tomato samples during survey. (A) One milligram (named 10┬░) total DNA of TYLCV-infected tomato or healthy tomato prepared from a commercial kit and the equivalent amount of crude sap from tomato infected with TYLCV (Ty) and healthy tomato (H) were serially diluted and subjected to the duplex PCR amplification. A relatively dilution points are shown on the top of gel. Sample prepared from different methods are indicated on the right. PCR products were separated on 1.2% agarose gel and stained with ethidium bromide solution. A 100 bp DNA ladder (M) was used as a size marker. DNA size was indicated on the left. (B) A representative agarose gel electrophosis of TYLCV detection using crude sap from tomato samples collected from various farms in different regions. As controls, crude sap from tomato infected with TYLCV (Ty) and healthy tomato (H) were simultaneously analyzed for the duplex PCR amplification of field-collected samples. lane 1-5 represents tomato cultivars susceptible to TYLCV and lane 6-10 indicates tomato cultivars resistant to TYLCV. PCR products were separated on 1.2% agarose gel and stained with ethidium bromide solution. A 100bp DNA ladder (M) was used as a size marker. DNA size was indicated on the left.
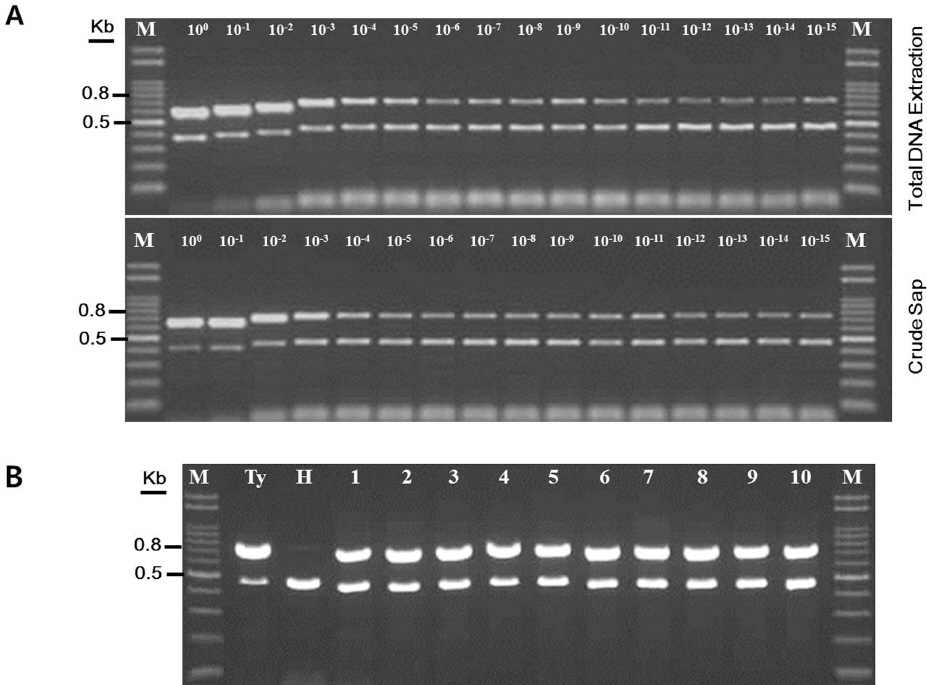
Next, we tested further efficiency of our duplex PCR method because tomato yellow leaf curl disease has been recently spread throughout the areas in which tomatoes produced. We investigated the incidence of TYLCV disease in 200 tomato samples from randomly selected 48 farms using the developed TYLCV detection method. PCR results showed that 72% tomato samples (144 tomato samples were positive for TYLCV) were infected with TYLCV (Fig. 3B and data not shown). Furthermore, some commercial tomato varieties (containing Ty-1 and Ty-3 genes) resistant to TYLCV were infected with TYLCV in the examined farms though the resistant tomato varieties didnŌĆÖt show any typical TYLCV symptoms in systemic leaves. Accumulations of TYLCV were lower in the resistant tomato varieties (data not shown). Results from the survey of tomato yellow leaf curl disease suggest that our duplex PCR method is less time-consuming and is suitable for large-scale survey on TYLCV detections, compared with conventional PCR methods.
In the present study, new sets of PCR primers to detect TYLCV isolates was designed and tested in the present study. A versatile method using this duplex PCR was developed for simultaneous identification of TYLCV isolates. The pair of primers specifically amplified tomato ╬▓-tubulin gene as an internal PCR control was developed. Moreover, the duplex PCR assay was optimized to amplify directly a part of TYLCV genome from leaf extracts. We anticipate that newly developed TYLCV primers are able to amplify all the TYLCV Korean isolates. For specificity and sensitivity of TYLCV primers, many pairs of primers that have been used to detect TYLCV isolates were validated by facts that the pairs of primers annealed at the corresponding sequence regions of target genes specifically and the pairs of primers could be used for amplification of target gene product at an extremely low amount of tomato DNA (Di Martino et al., 1993; Mart├Łnez-Zubiaur et al., 1996; Nakhla et al., 1993; Navot et al., 1991; Rojas et al., 1992; Roye et al., 1999; Wyatt and Brown, 1996). A few reports demonstrated that a versatile PCR protocol, using primer pairs, can be considered a good tool for detection of tomato yellow leaf curl disease in a single step. However, this method is not confirmed, when mixed virus infections or related TYLCV isolates are present. Previously, it was shown that TYLCV DNA molecule can be detected in tomato tissues and in whiteflies squashed on a membrane, followed by nucleic acid hybridization (Navot et al., 1989; Zilberstein et al., 1989) or PCR (Atzmon et al., 1998). The detection of TYLCV from an infected plant printed directly on Whatman 3MM paper is also possible (Navas-Castillo et al., 1998). A similar procedure was also used to amplify TYLCV directly from infected leaf extract. However, the addition of bovine serum albumin was used in the PCR reaction (Navas-Castillo et al., 1998).
The satisfactory results obtained from crude sap in our duplex PCR method may be explained by a low amount of inhibitors of the Taq DNA polymerase in the tomato tissues (Innis and Gelfand, 1990). The results reported here represent a significant improvement in conventional PCR procedures used to detect TYLCV isolates. Moreover, the use of PCR methods in conjunction with the minimal preparation procedure is proposed as a tool for quickly and easily classifying new isolates in combination with sequencing analysis. Thus, our duplex PCR method can be used in the laboratory as a tool for rapid and large-scale TYLCV diagnosis, since a small piece of tomato tissue is sufficient for detection.
In conclusion, this duplex PCR method allows with a high degree of accuracy and reliability for TYLCV detection.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print